Construction of ModelBricks
The original model
The creation of ModelBricks begins with a full model. As an example, we can take this model of a Rho GTPase cycle, which was published by Ota et al. in 2015 (PMID: 25628036).
|
|
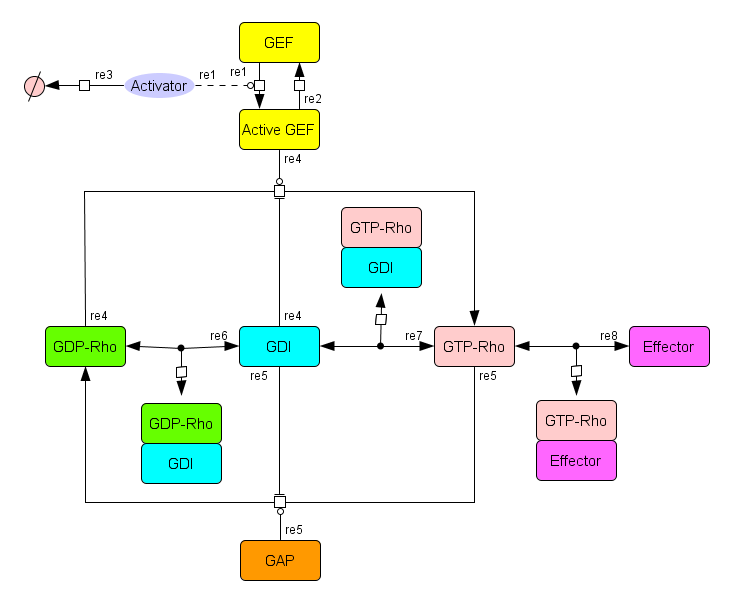
SBGN visualization
Figure 1B from publication |
In this publication, the authors are investigating possible positive regulation of Rho GTPase activity by RhoGDIs.
- Rho family GTPases: Act as molecular switches, controlling processes including actin cytoskeletal organization, microtubule dynamics, and vesicle trafficking
- Guanine nucleotide exchange factors (GEFs): Activate small G proteins by promoting the exchange of GDP for GTP
- GTPase activating proteins (GAPs): Inactivate small G proteins by promoting GTP hydrolysis
- GDP-dissociation inhibitors (GDIs): Negative role is to suppress overall Rho activity by inhibiting GEFs, while positive role is to sustain Rho activation by inhibiting GAPs.
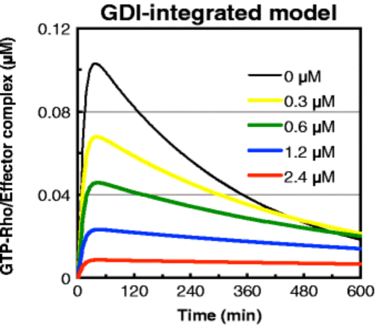
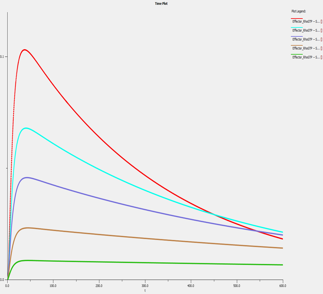
The SBML file provided in the publication was first imported into VCell. Simulations were run in VCell and confirmed to replicate those from the publication. As the concentration of intracellular free GDIs increases, Rho activation is prolonged, despite the overall decrease in Rho activity over time.
|
|
|
Simulation: Publication
Figure 2B |
VCell: Simulation |
Annotating the model
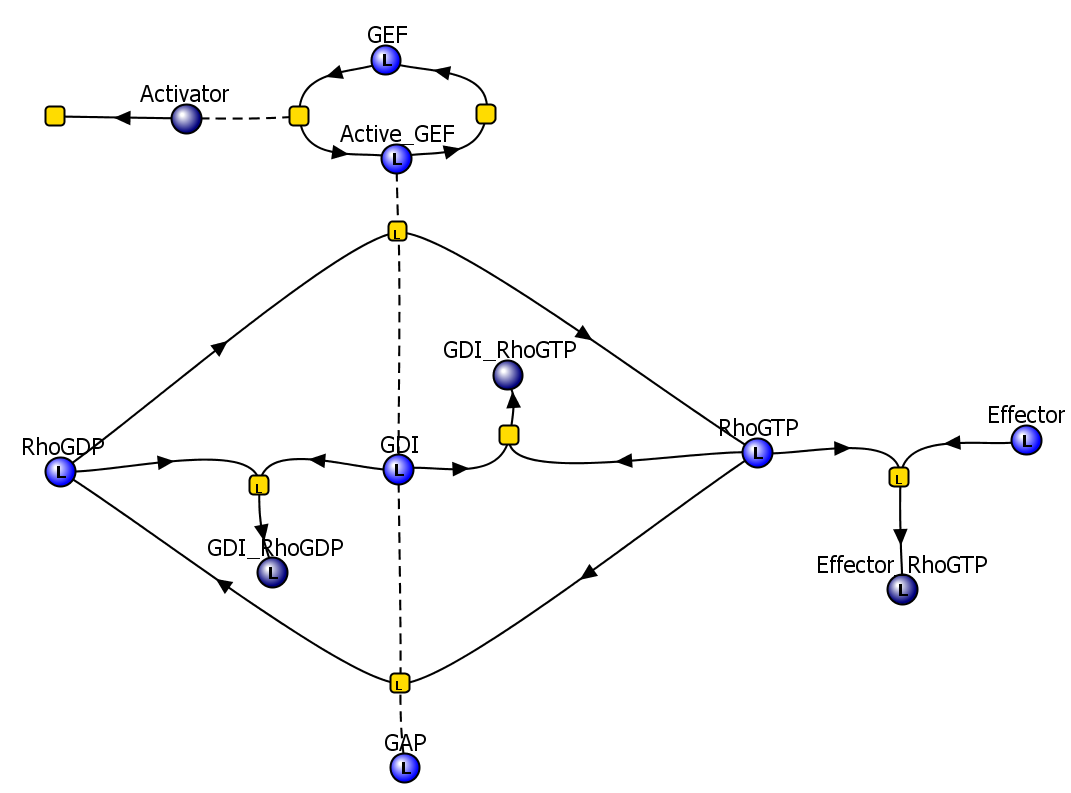
|
| VCell reaction diagram |
The model was then fully annotated to give the viewer a more thorough understanding of the model. Both textual and database annotations were added.
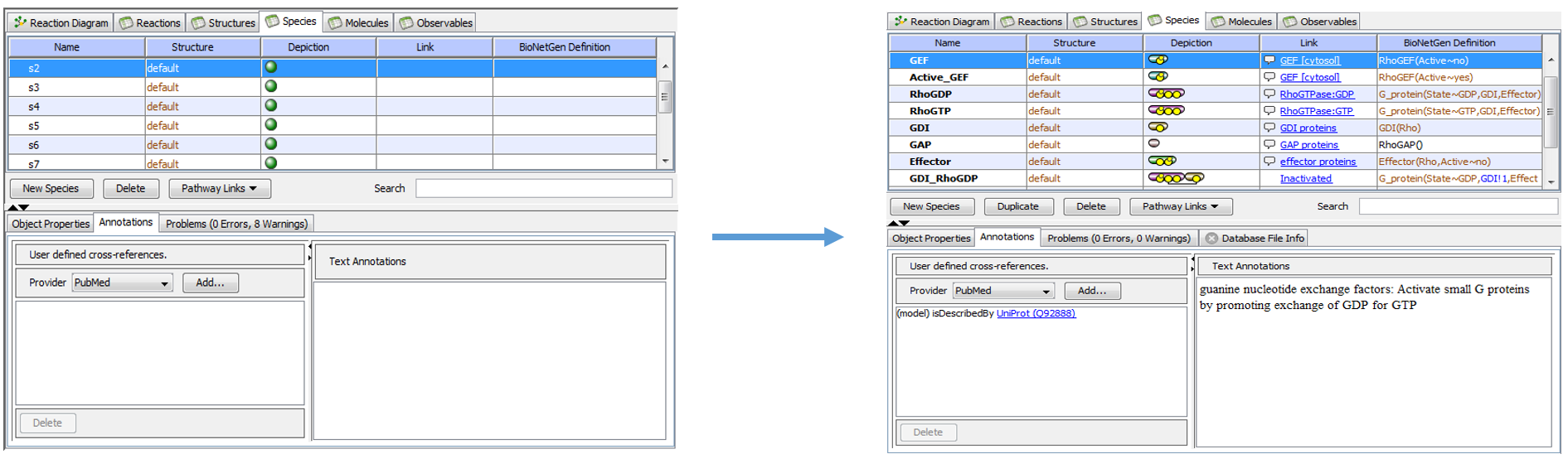
|
| Annotation of original model in VCell |
Species were modified so that they were constructed of individual molecules. By completing this step, each species became much more comprehensive and the constituent parts were clearly displayed.
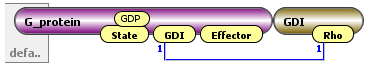
|
| Species composed of individual molecules |
ModelBrick creation
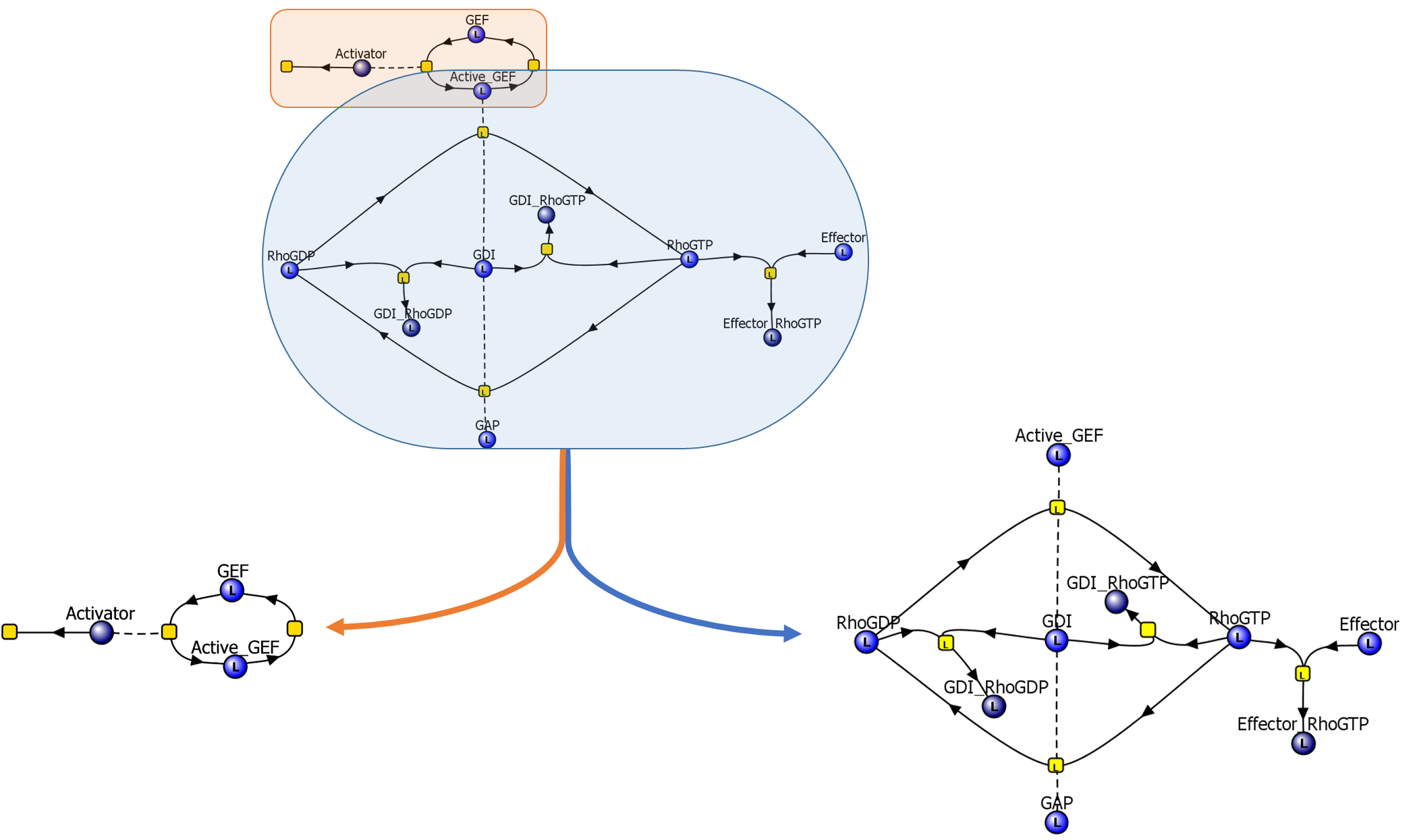
|
The first ModelBrick derived from the full model describes the activation of GEF and the degradation of the activator. The remaining portion of the model is its own ModelBrick, but it was further broken into two additional ModelBricks as well.
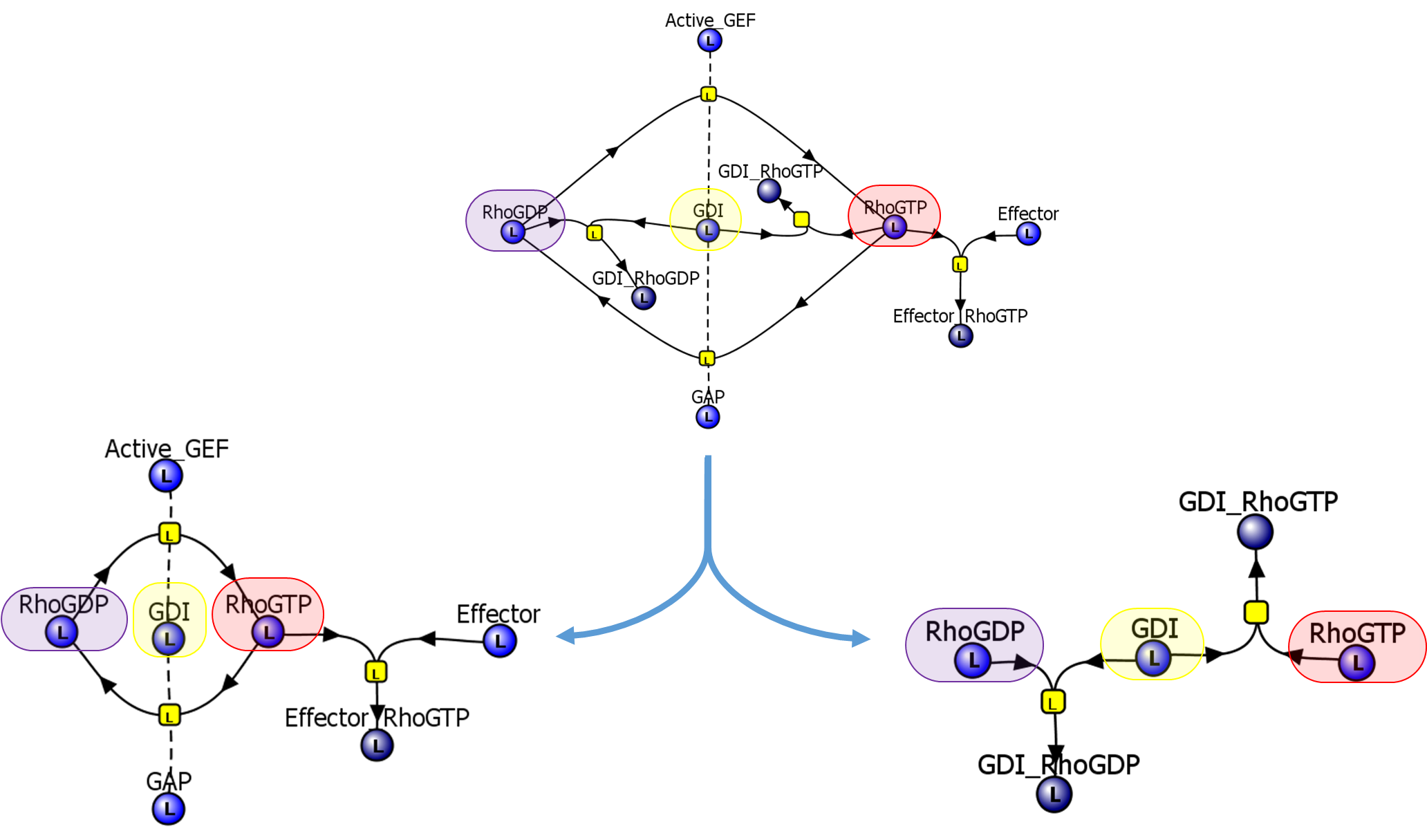
|
The two additional ModelBricks were created with one of them describing a much simpler Rho GTPase cycle and the other describing complex formation with GDIs and G proteins.
All ModelBricks are independent entities that can be combined to construct full models that are fully annotated and computable.